Author: Frank Dullweber, Böhringer Ingelheim / April 2016
Gene editing technology CRISPR-Cas
Modern biological research deals with approaches for modifying genetic information (genotype), i.e. the genetic makeup of a cell - and therefore of an organism. It is this information that determines the characteristics of that cell. In other words the change of the genotype can lead to an observable change of a cell or whole organism (phenotype).
Human diseases are based on different causes - from a clearly identified (mono) genetic disorder to influences of the environment as well as life style or a mixture of both. In pharmaceutical drug development it is becoming more and more important to understand the molecular basis of a disease in order to be able to modify this disease in an efficient manner. Using the vocabulary introduced in the first paragraph, we could say that the researcher who observes a specific phenotype would like to understand the underlying genotype to find the best medication for a patient.
A technique to study such relationships is gene editing, It can be seen as the molecular scissors to insert, delete or replace genomic information in the genome of a cell. The CRISPR-Cas technology is a relatively new, simple technology in the field of gene editing and was selected by the Science Magazine as the "2015 Breakthrough of the Year". This technique was initially described by Jennifer Doudna and Emmanuelle Charpentier. Several scientists call it the biggest biotechnological advance since the polymerase chain reaction (PCR). Its inventor Kary Mullis was honored with the Nobel Prize in Chemistry in the year 1993.
The CRISPR-Cas technology is of high interest in the scientific community. Therefore my goal was to identify the most important authors in this area by more objective indicators and guide colleagues to identify scientific conferences with speakers of high scientific expertise.
PubMed as a public resource
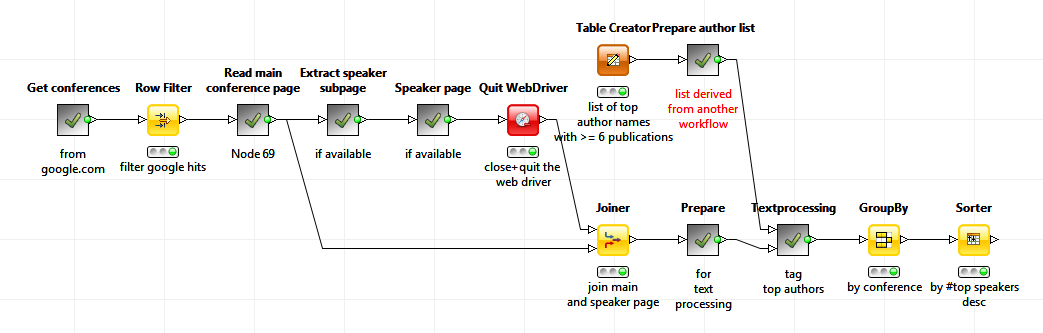
I used KNIME Analytics platform to collect, transform and integrate data from various internet resources. The whole process starts with a query against the PubMed/MEDLINE database.
This is a publicly available database of abstracts on life sciences and (bio-)medical topics. There are, in principle, two ways to access the PubMed database. The elegant way is the use of the Pubmed Query node that comes along with the Text Processing extension. For simplicity I just downloaded the answer set from PubMed and loaded the result via the File Reader node, which is part of the main KNIME installation.
Multivariate author ranking
One entry within the answer set is a publication in a scientific journal. Because I wanted to rank the authors of such publications, the results had to be transformed so that the author, identified by last name and the initials of the first name, becomes the grouping column. Once this transformation was done it was possible to generate the first attributes with the GroupBy node, e.g. number of publications, year of first and last publication, etc.
To further enrich the author with additional attributes I used the citation counts from the Scopus database from Elsevier to determine the mean numbers of citations as well as the author's h-Index. Last but not least, Thomson Reuter's famous list of Impact Factors (IF) for scientific journals was used to assess the importance of a specific scientific journal - the higher the citation rate per article, the higher the IF - although the sense of the IF has often been criticized. Both are commercial databases that need to be licensed by the user.
For now I had a list of authors enriched with several attributes like number of publications, mean IF or mean number of citations. This list was then used for multivariate ranking. Depending on your personal opinion about the more important or less important attributes you are able to change the ranking of the authors.
CRISPR-Cas9 is a customizable tool that lets scientists cut and insert small pieces of DNA at precise areas along a DNA strand.
Network visualization
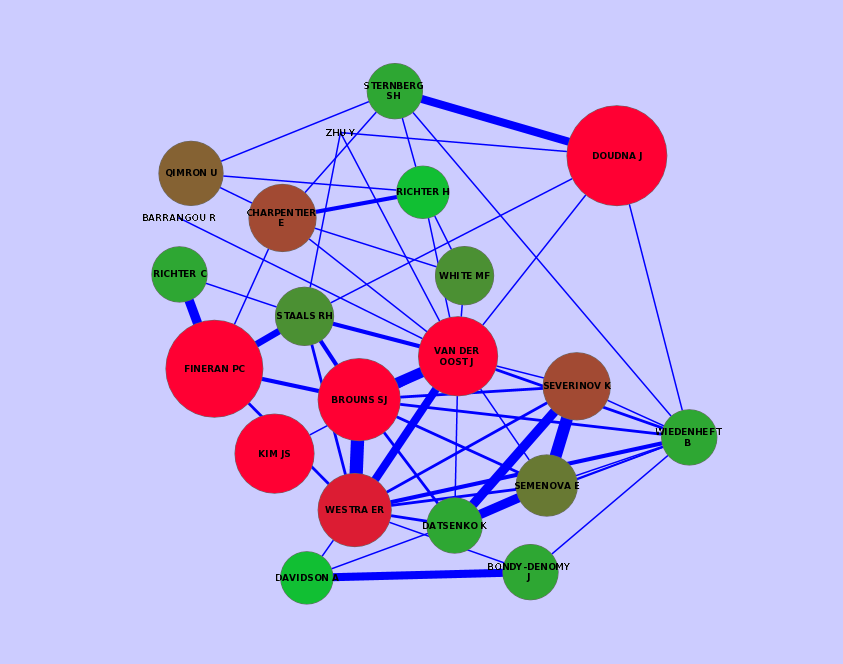
I also built the network of all the authors based on the transformation process of the PubMed data. With the help of KNIME's Network Mining extension I was able to construct and visualize the author network. There are several tools available to visualize the network. I typically use Cytoscape of version 2.x because there is a KNIME connector for this version.
Visualization of a subnetwork. The size and color stands for the number of publications for a specific author. The width of the edges describes the number of common publication for two authors
Author disambiguation
You may now ask if the ambiguity of author names can lead to false results and how this can be solved. A detailed solution to solve this problem completely is out of scope of my KNIME workflow, but I was able to improve disambiguation with some simple workflows. This was a multi-stage process that I will now try to explain in principle.
There were last names that existed with different but very similar initials (e.g. DOUDNA JA vs. DOUDNA J). This is likely to be the same person. More frequent names (e.g. Smith) or last names that exist more than twice with different initials were grouped together with their addresses based on their latest publication. This should be seen as an additional curation step because the number of publications and authors in our answer set allowed me to manually curate these groupings of authors. Please remember that I wanted to find the most interesting authors, therefore I did not need to take into account single authors with only a few publications.
Web crawling for conferences
I assumed that conferences with a lot of speakers from my ranked list of authors would be interesting to visit. The ranked list of authors could therefore be used to find interesting conferences. I used the new Selenium nodes for web crawling. First of all web address of conferences are collected via a Google search. Then the page content of the main conference pages as well as the content of pages about speakers and the conference agendas were downloaded and analyzed against the author list.
Summary
I have described how to extract and transform information with several publicly available and commercial resources. This information was used for a multivariate ranking of top authors. The generated ranking list was useful for visualizing author networks and also for searching for interesting conferences with well-known speakers.
Grouping literature based on authors helps us to understand different views on the same scientific topic. This is – for sure - a simple approach that can be further improved. But it adds some objective measures to assess conferences and to make an informed decision about participation in conferences.
Requirements
- KNIME 2.x or 3.x
- Extensions: XML Reader, Text Processing, Network Mining, Selenium (webcrawling nodes) together with PhantomJS executable
- Third party software: Cytoscape 2.x with extension KNIME Connector for visualization of networks (The connector can be downloaded from Cytoscape’s app store.)
