Reproducibility in science is coming under ever growing scrutiny. Datasets are becoming ever more diverse and complex. So doing reproducible science is getting harder, right? Well maybe not. KNIME gets reproducibility. KNIME gets diverse and complex. KNIME gets easy!
I am the new Application Scientist at KNIME. In my previous incarnation as a research scientist in Biophysics and Bioinformatics I was working as a Bioinformatics research scientist on the Virtual Liver Network, a major systems biology project in Germany involving some 70 research groups and 200+ research scientists. Along the way I learnt a few things, and I’m excited to have the opportunity to help the KNIME community to leverage the power of KNIME to make your lives easier.
Naturally one of the largest tasks within the project was managing the research data output. The team responsible for data management at HITS did an incredible job, helping scientists to understand why it is important to make their data available to other scientists. Even more important they were on hand to help the scientists to get their data uploaded into a data management system.
Automation
One major pain point for data managers in academic research projects are getting project members to upload their data. But what about building the upload to data management software into the research process? OpenBIS for example can be hooked up directly to an instrument in the lab, conveniently capturing all of the data (and metadata) from an experiment.
Now comes the cool part, the OpenBIS KNIME nodes can then allow users to build workflows to get their data out of the data store, and allow them to analyse their data with the most appropriate tools for the job. Know some Python, use the Python integration nodes. Know some R, use the R integration nodes. Don’t know a programming language, use the native KNIME nodes. Need to access external data sources like ChEMBL or the PDB, our community partners have built nodes for that. Your favourite web service doesn’t have a corresponding KNIME node, use the generic KNIME REST web services.
Someone in your lab made a cool workflow that you want to let other lab members use? Collapse the code into a metanode, and share using KNIME Server. Even better, what about KNIME workflows as scheduled jobs running on KNIME server? Facilities managers can get automatically generated usage statistics on a weekly basis. Principal investigators could have daily, weekly or monthly digests of an analysis using the most current data.
What science can KNIME do?
KNIME has trusted community contributions that means KNIME can analyse data from imaging (KNIP, HCS Tools), cheminformatics (CDK, EMBL-EBI, Erl Wood, RDKit, Vernalis), bioinformatics (NGS, OpenMS, openBIS). Additionally, there are many other nodes that have been contributed to the community that haven’t yet made it into the trusted contributions. If there aren’t nodes for your specific task, you can easily make your own!
A picture tells a thousand words
But what can scientists do with other scientists published data? Unfortunately, often nothing. Often the reason is that the data is not well enough annotated to be reused.
Not what you want to happen to your data…

The beauty of a KNIME workflow is that it naturally starts to produce the documentation for the analyses. Furthermore, it can easily document seemingly trivial data-processing steps, e.g. excluding a data point that is - obviously to the person who produced it, but not to the person trying to reproduce it - a mistake. Recently Phil Bourne and co-workers illustrated how much time it can take, and what might the limitations to reproducing a study be. Here the authors showed that it can take upwards of 280 hours to reproduce the analyses for a single research article. In this article one key point identified for producing reproducible research was to generate a flowchart that explains the steps taken in an analysis. Another point was to include all configuration files for an analysis. Well KNIME can take care of both of these steps!
Bundle your data with your analysis
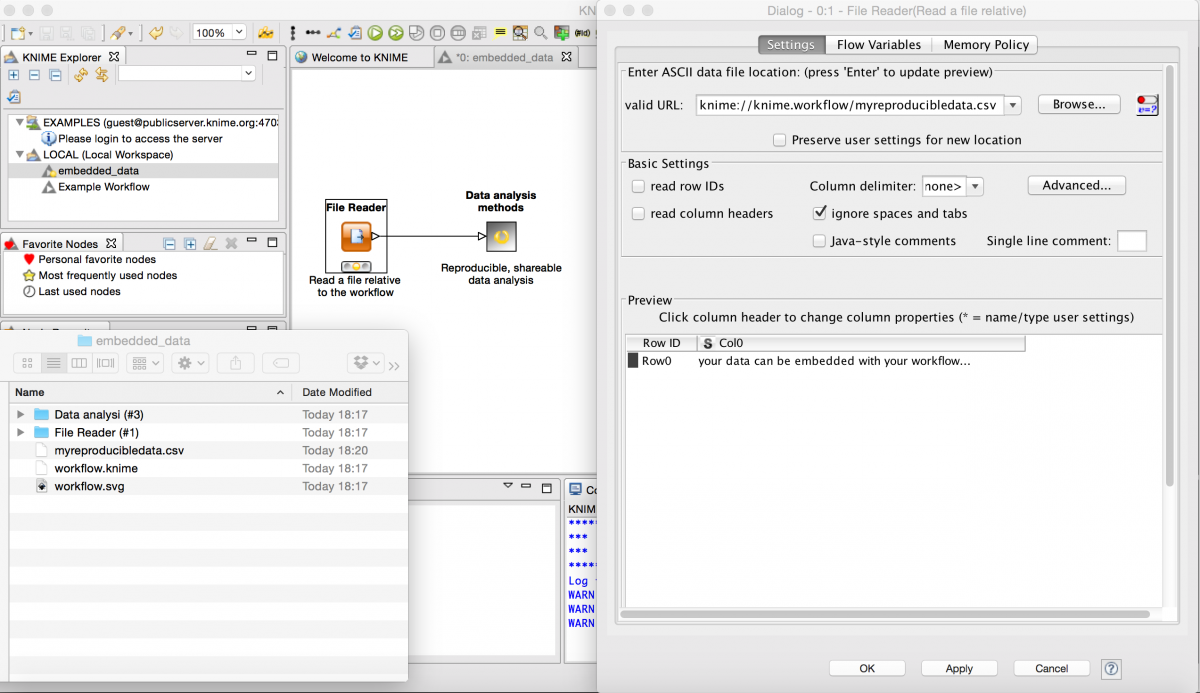
The workflow can be viewed as a flowchart that can be easily understood even by those from unrelated research areas, and additionally all of the configuration for the analysis is contained within the workflow. Even better, by using the KNIME protocol knime://knime.workflow/ followed by a relative path in the workflow directory, data files can be stored along with the workflow, meaning that the entire workflow is self-contained.
A self documenting workflow
Data-driven publishing
Being self-contained means that you could publish your research in an analysis-driven journal like GigaScience, where not only your research article is published, but also your data AND your analysis. In the case where you produced a cool dataset, but didn’t have the chance to do an analysis, you could publish the data in one of the new data-driven journals like Scientific Data. Here, KNIME could help to properly process the data before making it available to others, ensuring that error-prone manual data-transformation steps are automated and error free. Alternatively you could make your workflow available on a platform like figshare, where others can access, and also cite (through DOIs) your work. Even if you choose not to go down either of these routes, you could publish in a traditional journal and include your knime workflow as supporting information. All of these things need to, and are starting to happen in the academic world. At first it might seem like a lot of extra work, but with KNIME it can be a pleasure!
