Polycystic Ovary Syndrome (PCOS) affects as many as 5 million US women of reproductive age but getting a diagnosis isn't easy.
Research suggests that PCOS is associated with increased risk for diabetes. The disease is characterized by high insulin levels in the blood. This stimulates the production of more testosterone, which, in turn, can can lead to insulin resistance. Obesity is also a common feature of PCOS, affecting 40%-80% of patients. Insulin resistance and metabolic syndromes are common in obese women in general. The presence of both these medical conditions in a patient make it difficult to diagnose PCOS. Is it PCOS or obesity? Identifying powerful biomarkers could help resolve this problem.
Understanding the mechanism of a disease helps us understand what has gone wrong and identify what to target to treat the disease. However, clinical studies are frequently limited to a low number of people. Researchers can only work with small sample sizes, which fall short of reaching the necessary statistical power to be confident of the results. The ability to reveal insight from small samples would be useful, particularly as small clinical studies can address research questions within shorter timespans, and can be conducted within a single clinical center.
Daniel Contaifer (research specialist in health sciences) collaborated with his colleagues Dayanjan Wijesinghe (Professor at Virginia Commonwealth University (VCU)) and Monther Alsultan (PhD candidate at VCU) to propose using data mining techniques to resolve the complications in diagnosing PCOS, because of their ability to reveal previously hidden correlated features despite small sample size studies.
Resolving Complications in Diagnosing Polycystic Ovary Syndrome (PCOS)
PCOS is the focus of Daniel Contaifer's research. He uses KNIME to study associations between biomarkers – the molecules indicating normal or abnormal processes in the body which can be a sign of an underlying condition – and PCOS.
In this project, the team focused on biomarkers such as glucose and insulin, which are associated with PCOS in obese women. Using a KNIME workflow they were able to identify patients with PCOS from a small sample size study.
In this article we want to walk through the workflow and show you how it can be used to:
-
Explore, oversample, combine, and process metabolic, lipidomic, and clinical data;
-
Build and optimize a Random Forest classifier to identify biomarkers that distinguish patients with PCOS from those without it;
-
Validate the relevance of these putative biomarkers.
Multivariate Analysis for Biomarker Exploration
The use of multivariate analysis helps explore putative biomarkers for diseases through the lenses of biochemical and metabolic imbalance. However, using multiple data sources to understand the mechanisms of a particular disease can fall short, due to the common limitations of studies with small sample sizes. Clinical studies are frequently limited by a low number of subjects, and consequently do not reach the desired statistical power to assure confidence in the results. Regardless, these studies can address research questions in a short timespan, and can be conducted in a single clinical center.
The authors propose that mining distinct datasets can reveal underlying correlated features, even with small cohorts. The workflow discussed here, built with KNIME in a completely codeless fashion, is a take on this proposal. It is used to find putative biomarkers to help identify patients with PCOS in a small sample size study.
Derive Insight from Limited Samples with Multivariate Analysis
This workflow is part of a bigger project defending multivariate analysis as a valid approach to tackling limited samples, which are very common in biomedical research. The idea is that even if a study does not include enough samples for a statistically significant analysis, insights can still be derived with the help of multivariate analysis. This in turn can lead to data-driven hypotheses and help test biomedical strategies. Daniel used a Random Forest predictive model to uncover biomarkers that help distinguish obese women with PCOS from those without. These parameters were then used for testing their power as putative biomarkers.
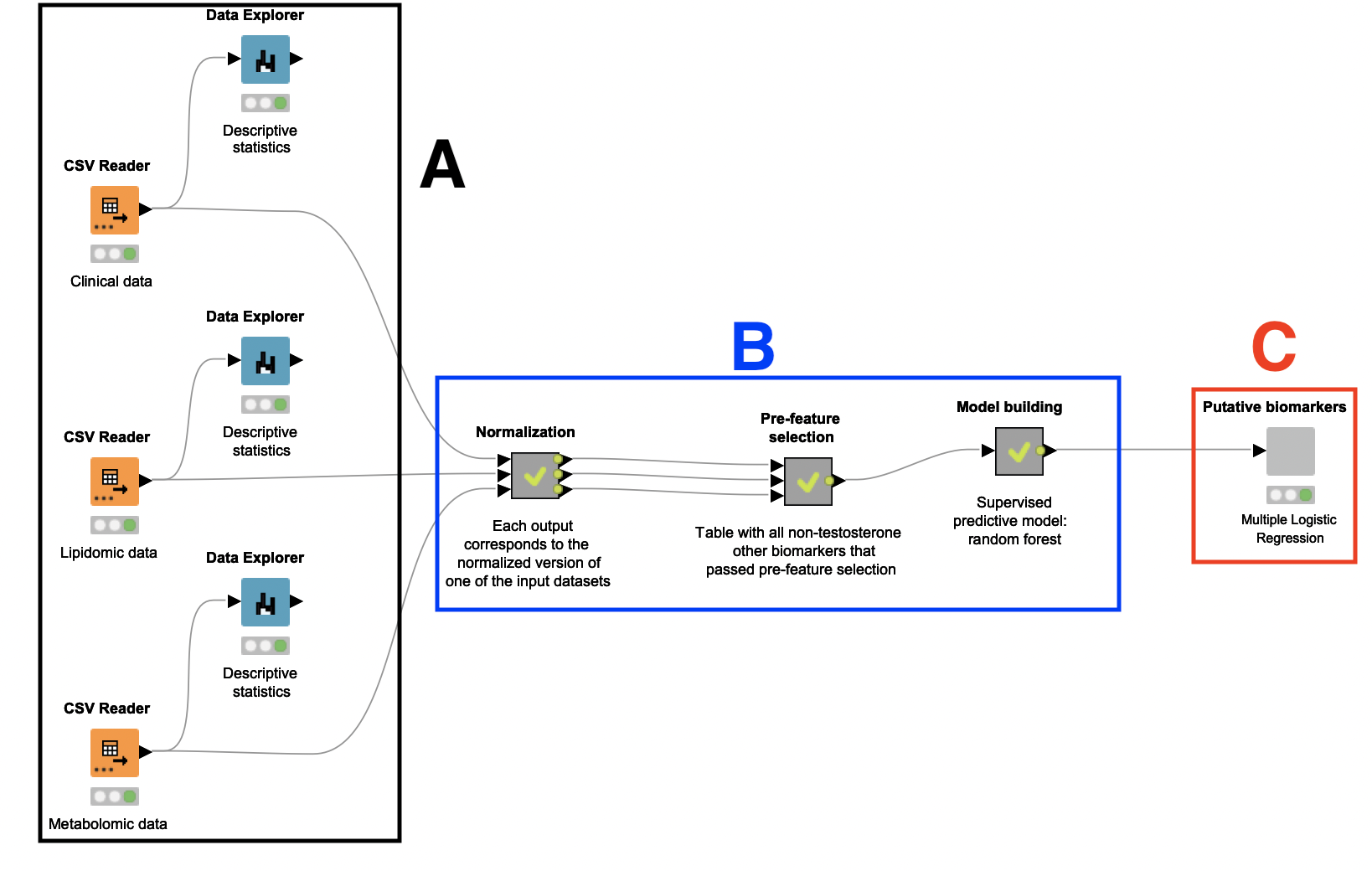
A: Dataset selection and descriptive analytics
This study relies on a small number of patients: 15 women who are diagnosed with PCOS, and 15 women who are not. Their plasma samples were collected, anonymized, and analyzed, and the resulting clinical, lipidomic, and metabolomic datasets were used as input for this workflow. These datasets contain a variety of biological parameters that may be correlated with PCOS, such as BMI, insulin level, and relative levels of different lipids and amino acids. There is also an attribute indicating whether the subject has PCOS.
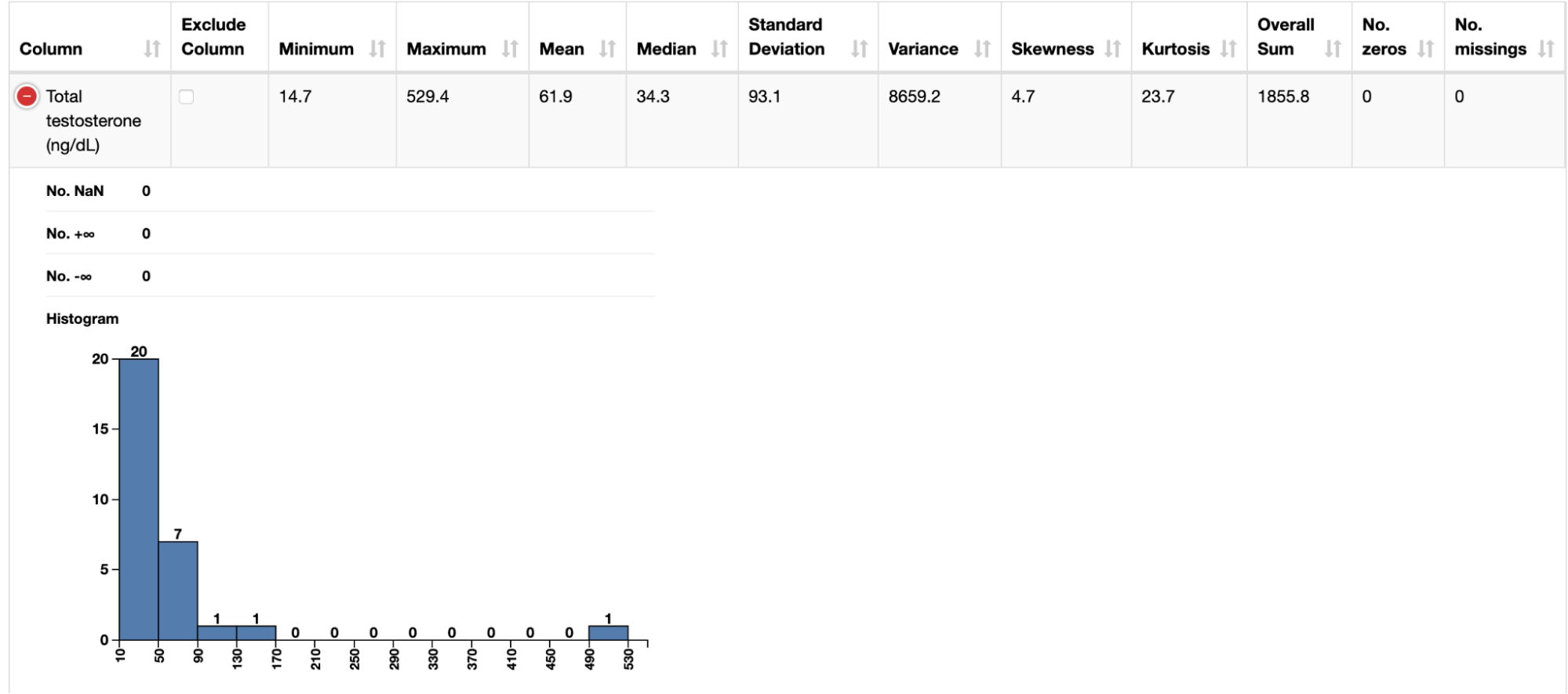
After ingesting these datasets with instances of the CSV Reader node, we connected each of them to a Data Explorer node to facilitate inspection of their attributes. Through a JavaScript view, it is possible to quickly check how different biomarkers (e.g. BMI and testosterone level) are distributed across the patients.
B: Data preprocessing, feature selection, and model creation
A Random Forest classifier is built to identify the attributes which best indicate a PCOS diagnosis. To build this classifier, it was necessary to first preprocess and oversample the datasets. A feature selection step was also performed to remove attributes that are likely not related to PCOS.
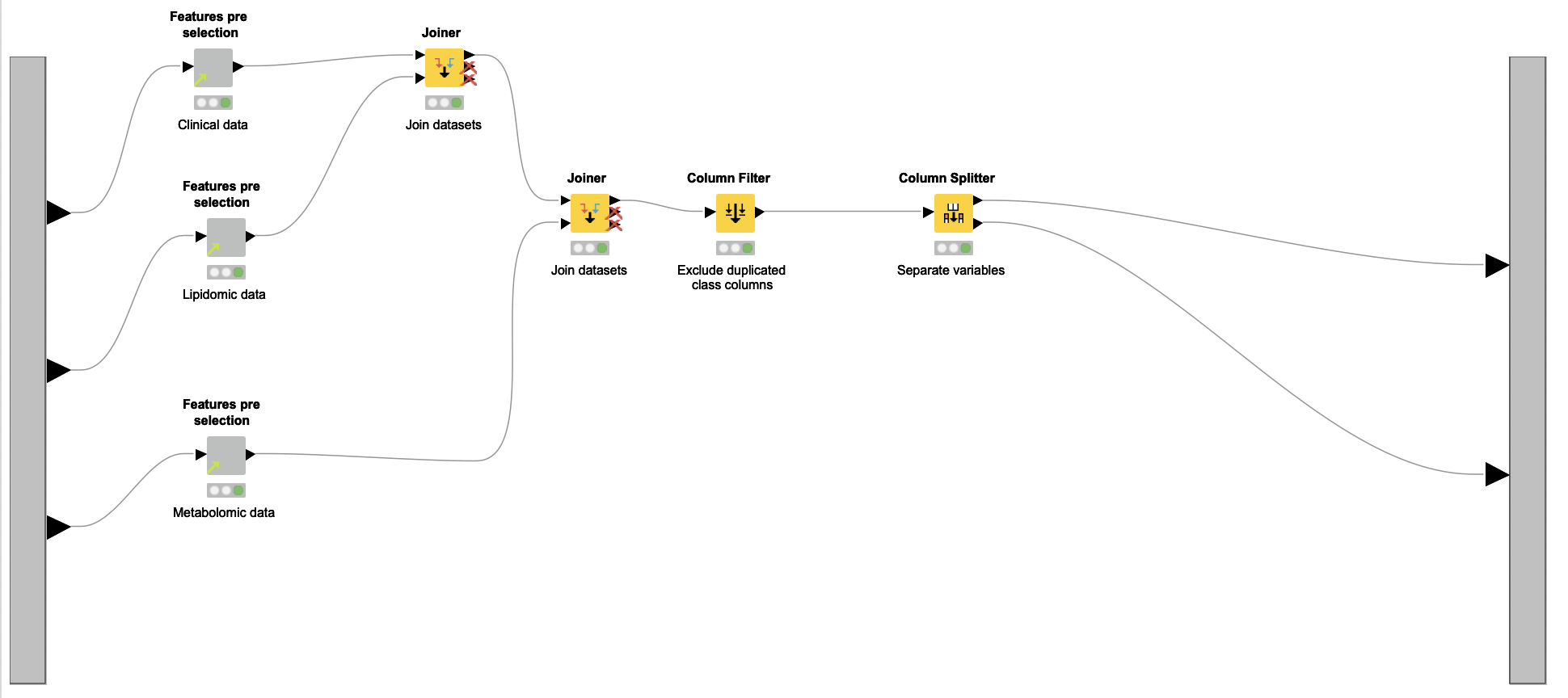
Data Preprocessing. With the datasets at hand, the next step is to preprocess them. To this end, the authors created a reusable shared component named Data Normalization, which is applied over each dataset independently inside a metanode named Normalization (see Figure 1, region B).
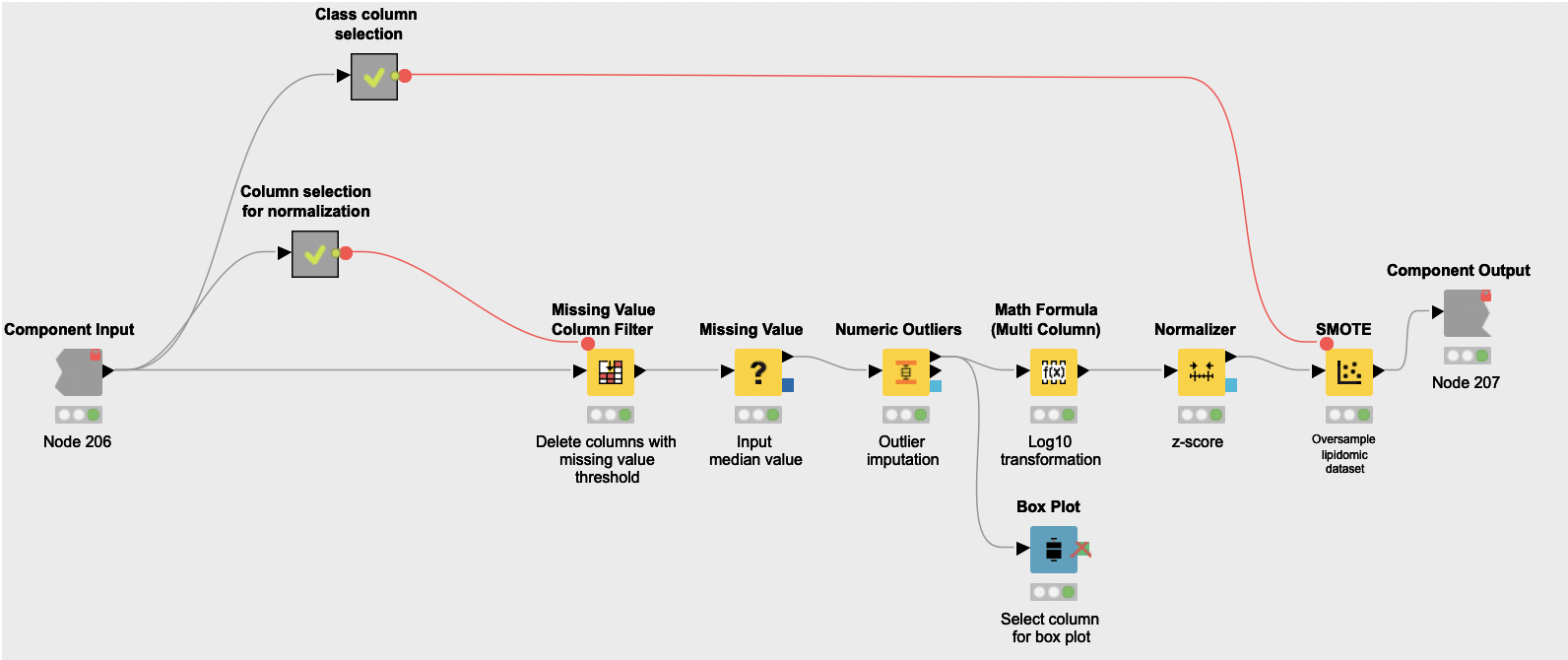
The shared component Data Normalization works in three steps:
-
First it addresses missing values and outliers. The Missing Value Column Filter node is used to remove attributes that have more than 15% of missing values, as they are likely to have quality issues and lead to a more imprecise analysis. Then the Missing Value node is used to replace empty values with the median of their corresponding attributes, and the Numeric Outliers node is used to identify and dampen outlying values.
-
Then the normalization actually happens. Numeric attributes from all datasets end up represented in the same scale to facilitate posterior interpretation. First a logarithmic transformation of the datasets (Math Formula (Multi Column) node) spreads values in such a way that they get closer to being normally distributed. These new values are then turned into z-scores with the Normalizer node.
-
Finally, the component oversamples the datasets with the SMOTE node, which creates synthetic rows by extrapolating between a real observation of a given class and one of its nearest neighbors of the same class. Note that while small sample sizes often sub-represent the overall population, this workflow operates under the assumption that this sample is representative of the variation that characterizes the pathophysiology of PCOS. Consequently, oversampling with SMOTE likely leads to a consistent training dataset for posterior classification models.
The output of metanode Normalization is the preprocessed version of the clinical, lipidomic, and metabolomic datasets. These datasets are cleaned, normalized, and ready for the next step of the analysis: feature selection.
Feature Selection. The feature selection process starts with the removal of features with very low variance (Low Variance Filter node) from all normalized datasets. The reasoning here is that such features are probably not related to PCOS and thus not very informative. Next, features that have a high linear correlation are eliminated (Linear Correlation and Correlation Filter nodes), due to their redundancy. Since the same process is applied over all datasets, Daniel encapsulated it in a shared component named Features pre selection. Once the process is done, all remaining unique features are aggregated (Joiner node and Column Filter node).
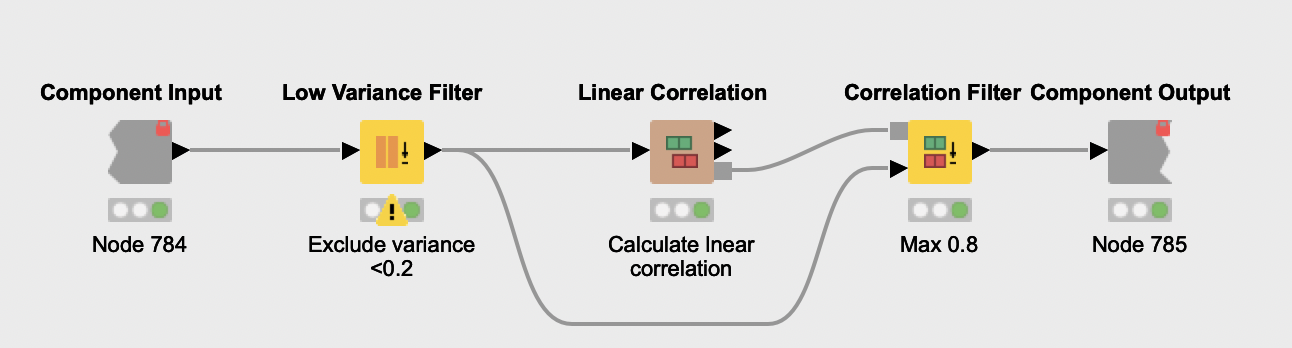
The Column Splitter node is then used to separate testosterone-related features from the other selected biomarkers. Only the latter are used to train a Random Forest classifier. The authors exclude testosterone indicators because they are already known as powerful biomarkers for PCOS. If used, they can hamper the discovery of other discriminating biomarkers.
The workflow segment responsible for this initial selection of biomarkers is encapsulated in the Pre-feature selection metanode.
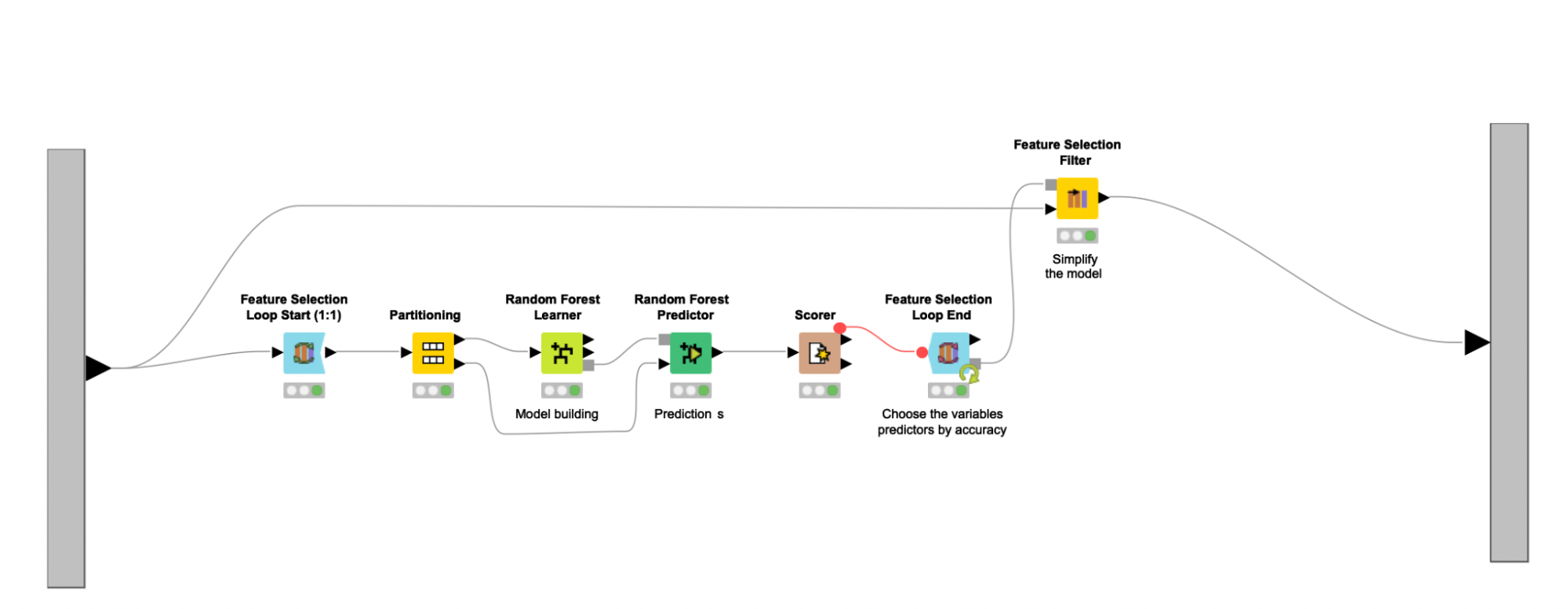
The feature selection process continues with the help of a feature selection loop (Feature Selection Loop nodes). Here the Random Forest classifier is trained on different feature sets, and then the set associated with the highest accuracy gets selected. The feature sets fed to the Random Forest Learner node are chosen randomly, and an upper bound of 16 is used to enforce that such sets are never too large, representing around 25% of the final combined dataset. Other feature selection methods, such as forward feature selection or backward feature elimination, were explored and led to similar results in terms of performance. Since random selection executes faster, the author settled with this strategy. Other classifiers (e.g., SVMs, Decision Trees) were also studied but did not perform as well as the Random Forest.
Note. If you want to re-use this workflow with your own data, Daniel advises exploring different classifiers.
After the feature selection loop executes, the Feature Selection Filter node is used to filter out all columns that are not a part of the best feature set. The selected feature set is assumed to represent the most important markers considered for the diagnosis of PCOS. Useful clinical information can also be derived from the selected feature set, related to the discovery of putative biomarkers and to the creation of data-driven hypothesis tests. This entire process is enclosed in the Model building metanode. The selected set of features suggest five clinical parameters, five lipids, and six metabolites as predictors characterizing the presence of PCOS in obese women.
C: Putative biomarker discovery
In the last part of the workflow, encapsulated in the component Putative biomarkers, the author verifies whether the predictors selected by the Random Forest classifier can be considered putative biomarkers. If so, these predictors should be further investigated as criteria that can characterize PCOS.
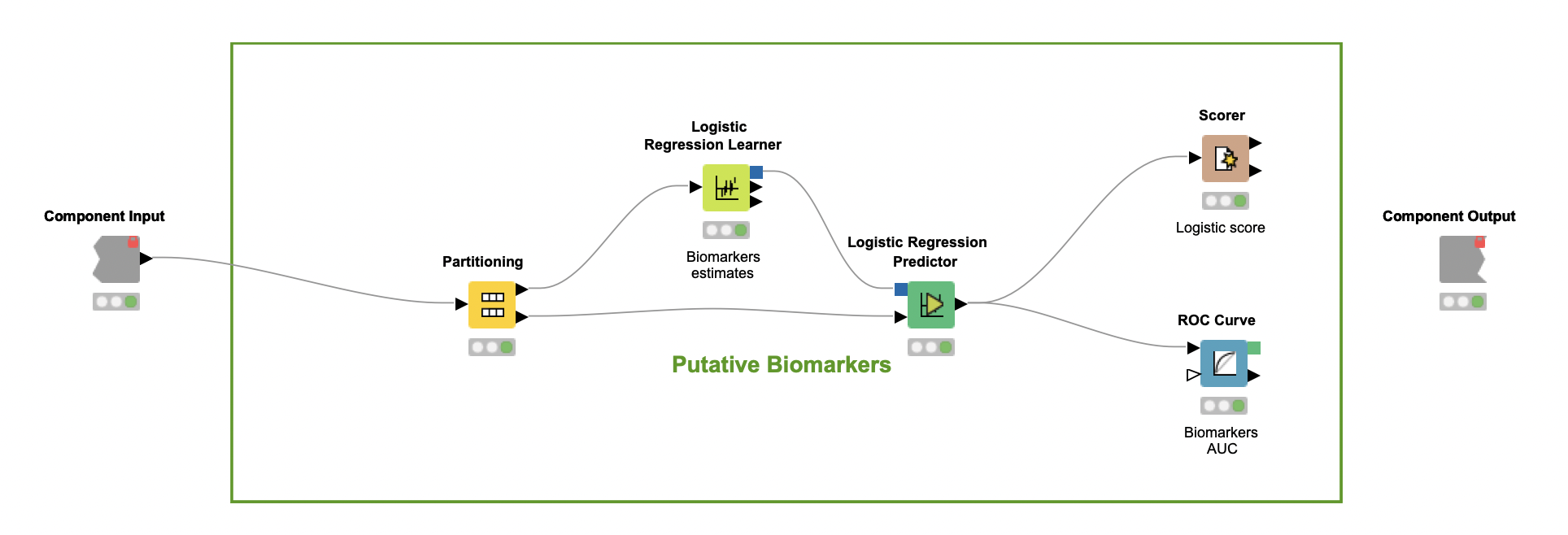
To carry out this investigation, a Logistic Regression (LR) classifier is trained over the set of selected predictors alone (Logistic Regression Learner node). This model, which assumes a linear relationship between multi-covariates and categorical classes, leads to an F-measure close to 83% for both classes (Scorer node). These numbers suggest that this combination of predictors may help characterize PCOS to a certain extent, but are not enough to uncover how each relates to the syndrome.
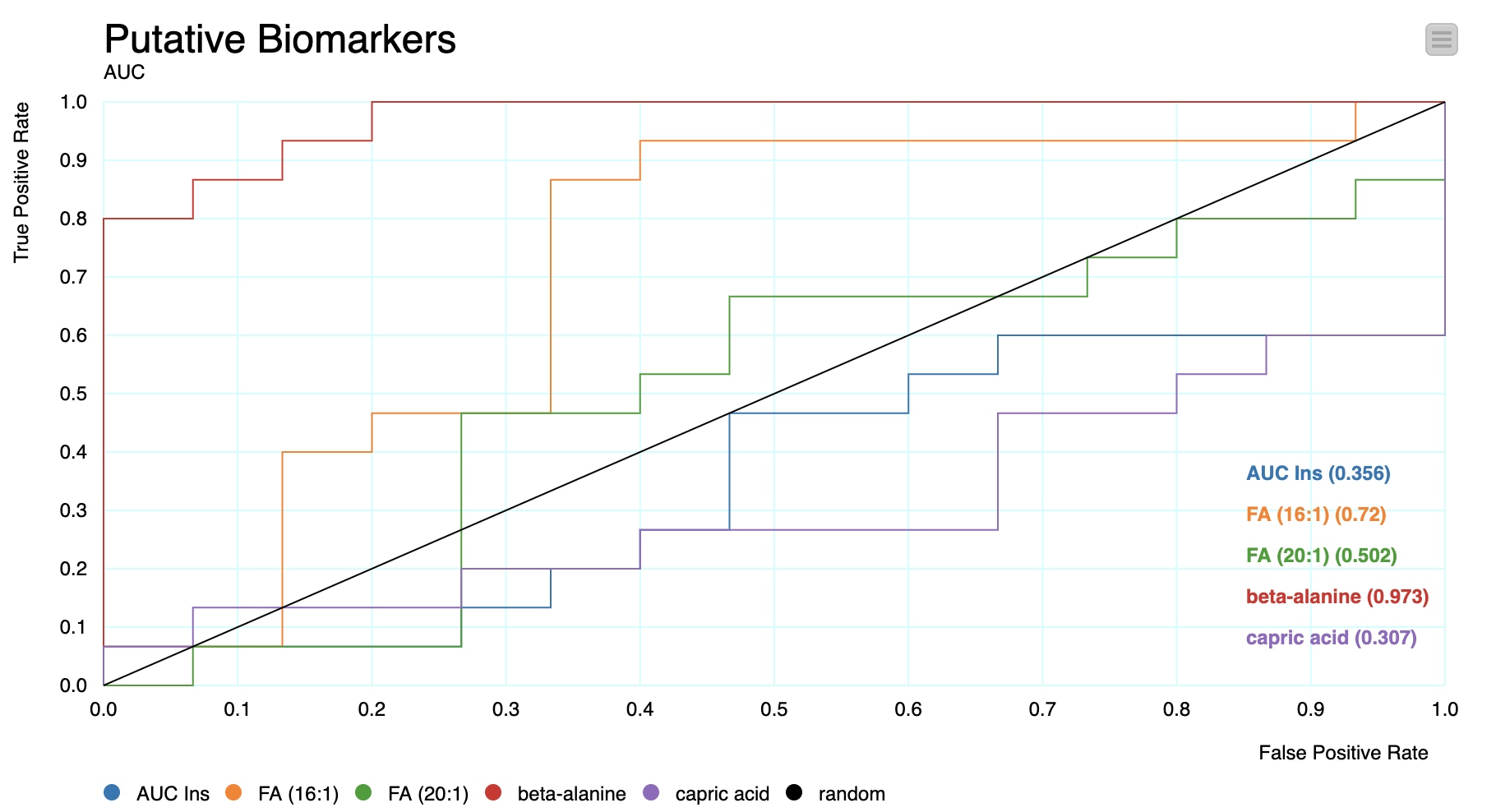
To better understand this, the Receiver Operating Characteristic (ROC) curve is used (ROC Curve node). For all possible values of the C-statistic, the ROC curve plots the sensitivity of each predictor against one minus its specificity.
Note. The C-statistics gives an estimation of the Area Under the Curve (AUC) of the ROC. In this case, each C-statistic value indicates whether a patient's chance of being classified as having PCOS is above the curve.
These values indicate whether different predictors have a strong and independent correlation with the outcomes – in such cases, they may have value as putative biomarkers. For the particular node configurations of this workflow, the best predictor not related to testosterone that was found was beta-alanine, an aminoacid.
A Powerful Workflow to Analyze Small Sample Size Clinical Study Data
The proposed multivariate analysis workflow — powerful, but simple to build with KNIME — is an interesting example of how to handle studies with small sample size studies. The transformation and normalization of the datasets allows data standardization for model building, and the oversampling strategy reinforces the training set while maintaining the variability of the original data. The option to use the original cohort as a test set improves the power of validation, overcoming the problem of small samples. Moreover, strong evidence of the relevance of selected predictors supports the usefulness of the workflow for exploration of putative biomarkers.
A statistical workflow that can be used for the reliable replication of results is strongly recommended when working with multiple datasets. In addition, the possibility of applying the principles of systemic biology allows the study to be used toward a more personalized medicine. This was exemplified with the PCOS study analysis, which revealed strong predictors representing different characteristics of the PCOS pathology. The opinion of the authors is that KNIME can be used to facilitate data preprocessing and analysis, and it also offers a clever oversampling strategy to overcome the problems with small samples
